the Stephen Floor lab
The Floor lab was open from 2017 to 2024. Stephen has moved to Inceptive.
Contact: stephen@floorlab.org
The Floor lab was open from 2017 to 2024. Stephen has moved to Inceptive.
Contact: stephen@floorlab.org
Our cells are like machines. Genes in our DNA are the instructions to produce each part (proteins, RNA, and other molecules). Each part must be accurately produced in the right amount for cells to work properly. Mistakes in these processes cause many diseases, including cancer.
Messenger RNA sequences instruct the cell what protein part to make and how much to make of it. Other RNA molecules also affect how cells work. Our lab studies how healthy cells make decisions and how decisions change in human diseases.
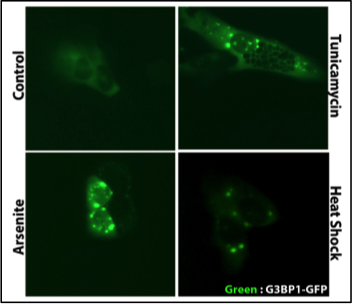
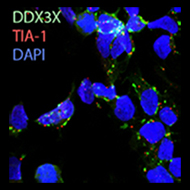
Left: phase separation like oil and water of RNA binding proteins and RNA into round foci under cellular stress.
We study the many ways RNA influences human gene expression. How do mRNA sequences specify protein production levels? How do changes to the biophysics of RNA in cells influence gene expression, and what role(s) do RNA chaperones play? How are these processes changed in human diseases, and can the changes be corrected?
The Floor lab uses genome editing, functional genomics, biochemistry, and cell biology to explore these and other questions. Learn more about our projects or recent work.
We are members of the Department of Cell and Tissue Biology & Helen Diller Family Comprehensive Cancer Center, and Biophysics, Biomedical Sciences, & Tetrad Graduate Programs.
Thanks to the UCSF PBBR, the California Tobacco Releated Disease Research Program, the NIH New Innovator program, the NIH/NINDS, the NIH/NIGMS, the Innovative Genomics Institute, the Pew Charitable Trusts, and the DDX3X Foundation for supporting our work!
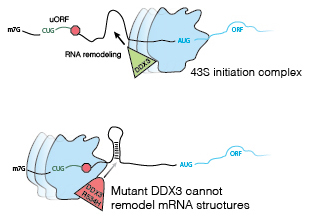
Remodeling of RNA by DEAD-box RNA chaperones
DEAD-box proteins use the energy of ATP hydrolysis to remodel RNA structures and RNA-protein complexes. We study how DEAD-box proteins influence gene expression through mechanical work.
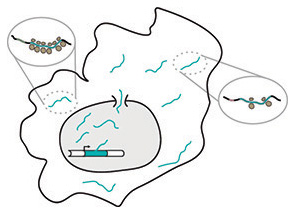
Translational output of the human transcriptome
Most human genes give rise to multiple mRNAs through alternative mRNA processing. Each mRNA may include different coding sequences, regulatory sequences, or both. We measure how well different mRNAs are translated and use this information to control transgene expression.
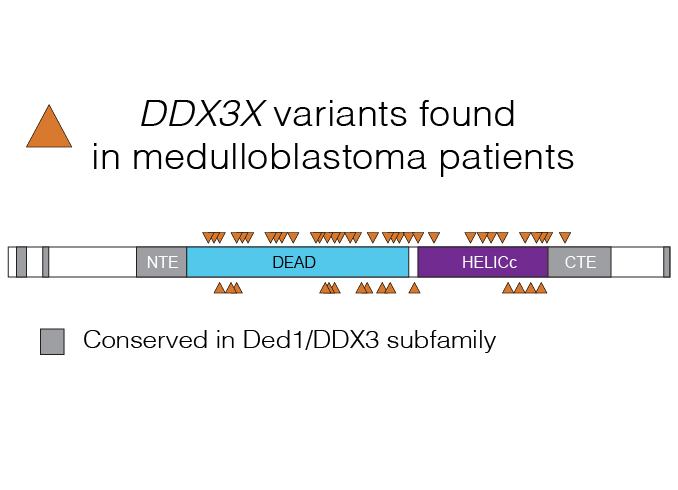
RNA in developmental disorders and cancer
DEAD-box proteins and other aspects of RNA biology are altered in numerous human diseases. We are studying how alteration of the DEAD-box protein DDX3 drives formation of the brain tumor medulloblastoma, and other RNA-related processes in disease.
Cell biophysics & phase separation of RNA
RNA molecules exist in a crowded cytoplasm. RNA and RNA-binding proteins partition into phase-separated RNA granules under some conditions. We investigate the role of RNA chaperones like DEAD-box proteins in this balance.
Or most of us anyway

Principal Investigator (CV)
stephen@floorlab.org
B.S. Physics, Computer Science: Univ. of Kansas
Ph.D.: UCSF Biophysics (John Gross lab)
Postdoc: Berkeley (Doudna lab)

Graduate Student (Tetrad)
B.S. Industrial Biotechnology:
University of Puerto Rico, Mayaguez
NIH/NIGMS F31 Fellow

Postdoc
B.S. Biology, University of Science and Technology of China
Ph.D.: Carnegie Mellon University Biological Sciences (Joel McManus lab)


Postdoc (joint with Ron Vale)
B.S.: Biochemistry: U. Rochester
PhD: Scripps Integrative Computational and Structural Biology
(Ian MacRae lab)

Postdoc
B.S.: Mol Cell & Dev Biology: U. Washington
PhD: UC Berkeley Molecular and Cell Biology
(Gloria Brar lab)
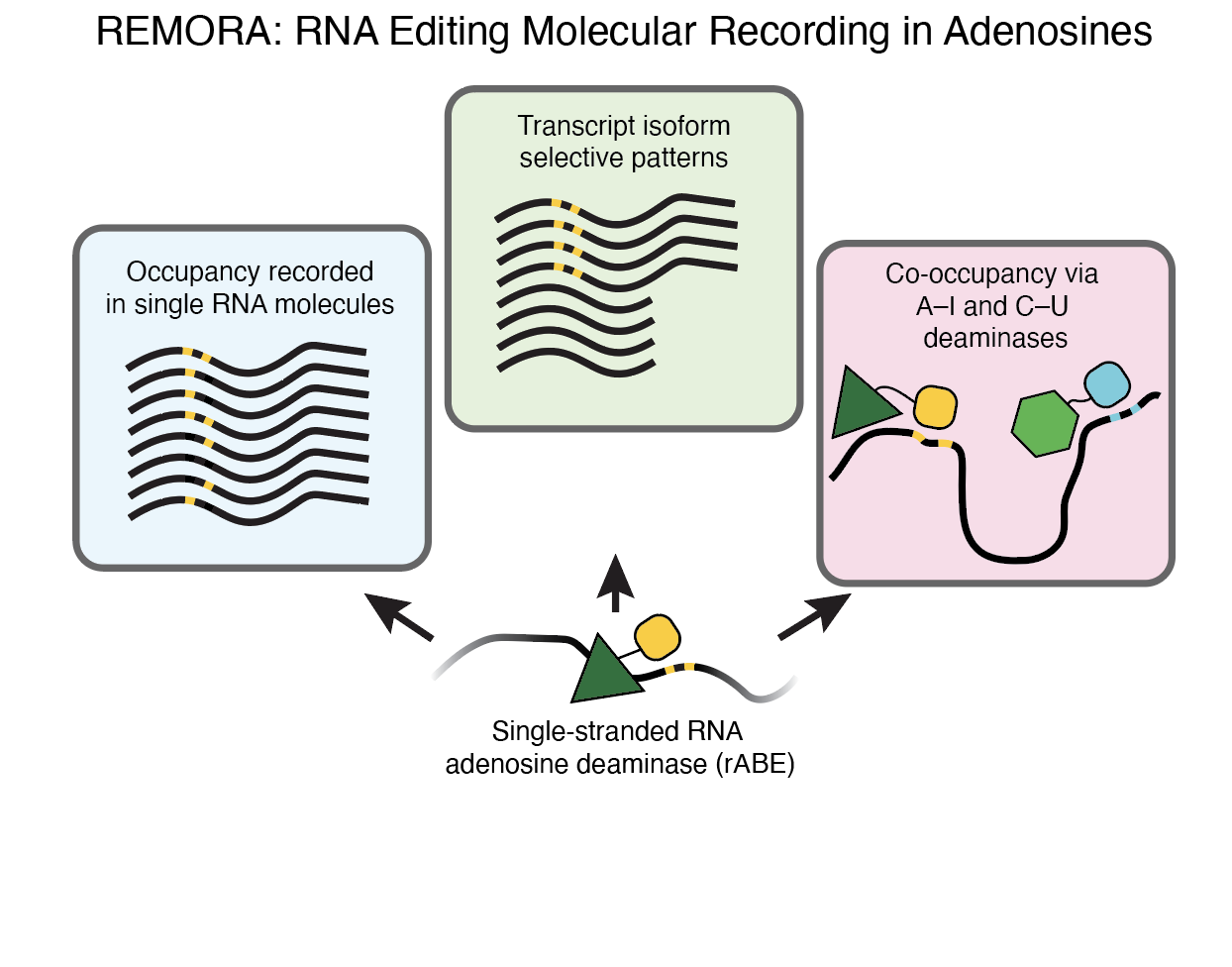 RNA molecular recording with an engineered RNA deaminase
RNA molecular recording with an engineered RNA deaminase
Lin Y, Kwok S, Hein A, Thai B, Alabi Y, Ostrowski S, Wu K, Floor SN. (2023)
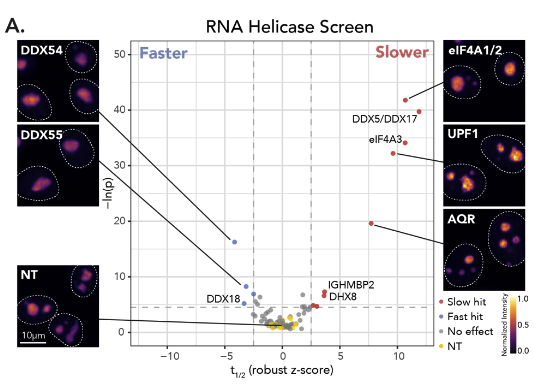 Nucleolar dynamics are determined by the ordered assembly of the ribosome
Nucleolar dynamics are determined by the ordered assembly of the ribosome
Sheu-Gruttadauria J, Yan X, Sturrman N, Floor SN, Vale RD. (2023)
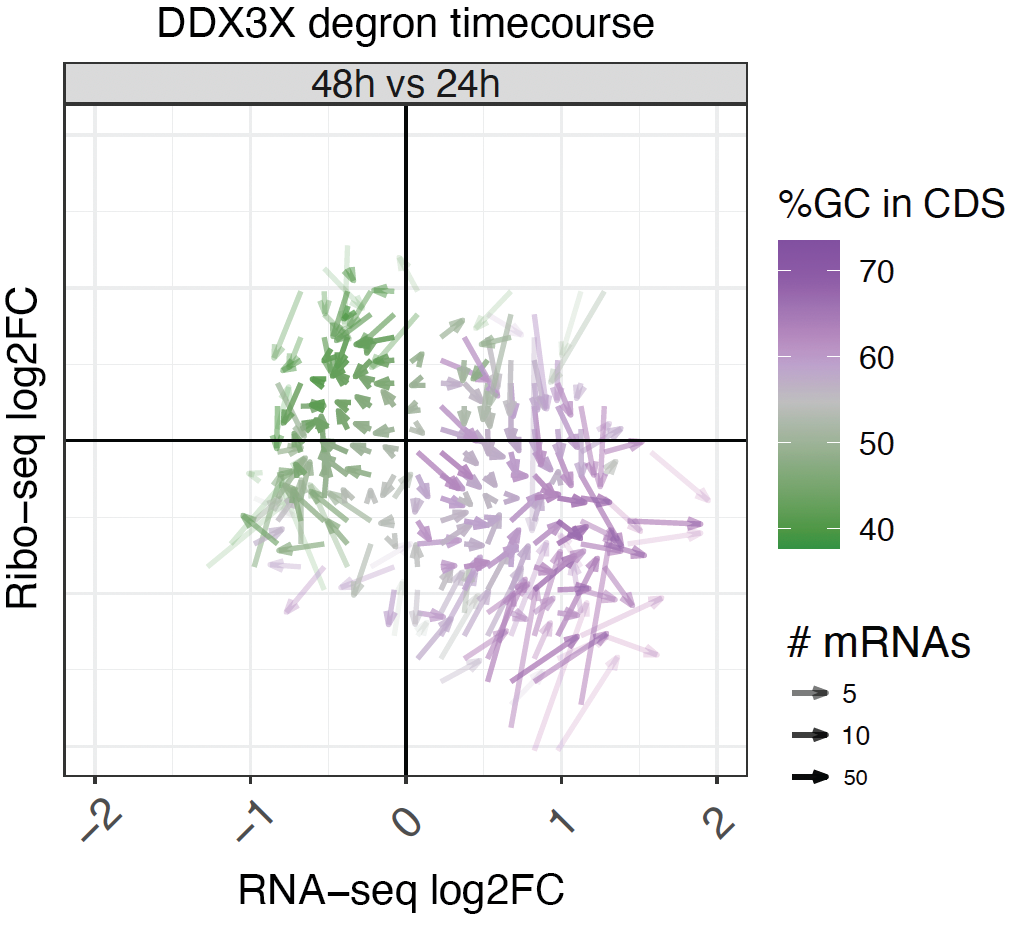 A ubiquitous GC content signature underlies multimodal mRNA regulation by DDX3X
A ubiquitous GC content signature underlies multimodal mRNA regulation by DDX3X
Jowhar Z, Xu A, Venkataramanan S, Hoye M, Silver D, Floor SN, Calviello L. (2023)
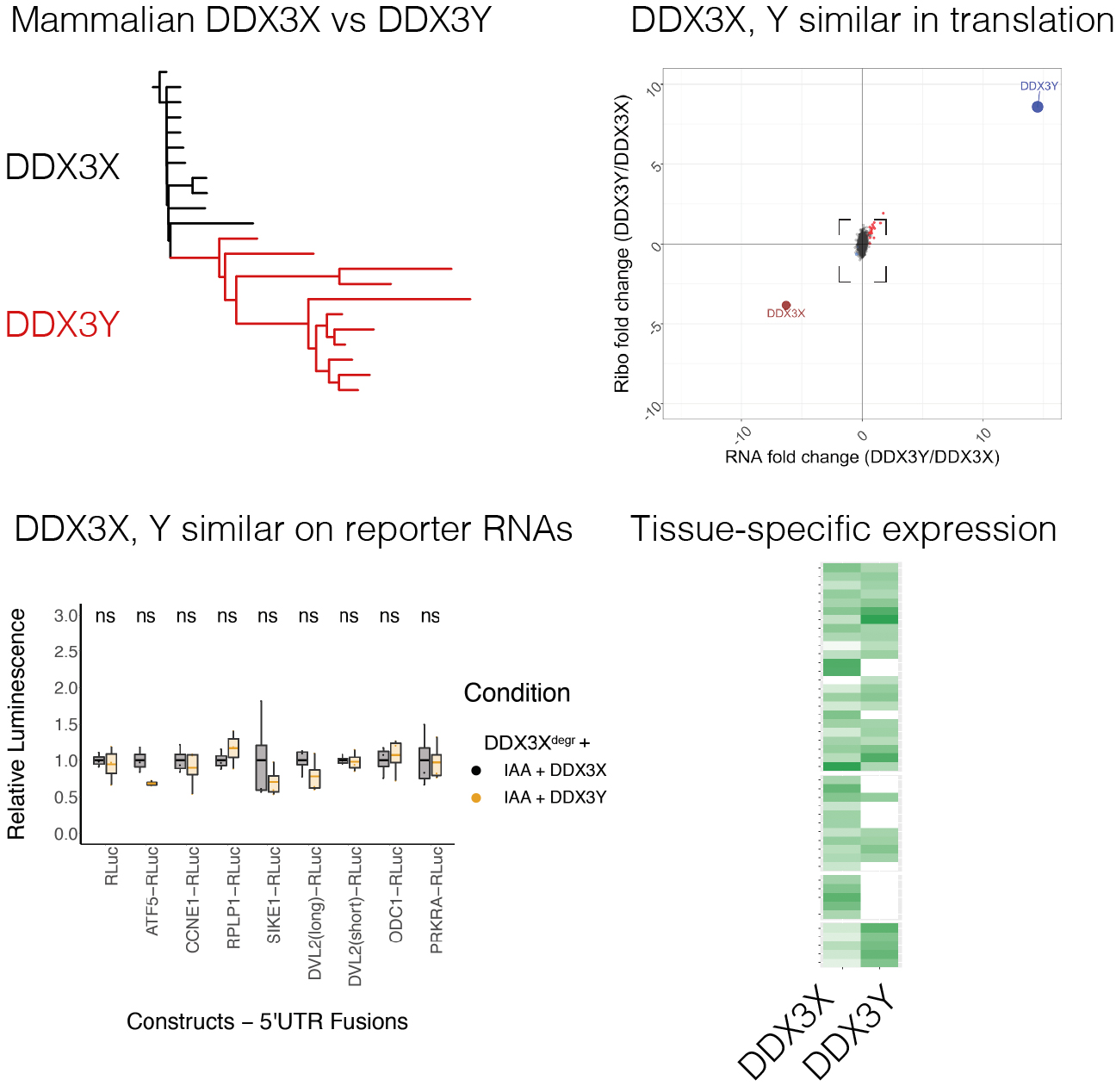 DDX3X and DDX3Y are redundant in protein synthesis
DDX3X and DDX3Y are redundant in protein synthesis
Venkataramanan S, Gadek M, Calviello L, Wilkins K, and Floor SN. (2021)
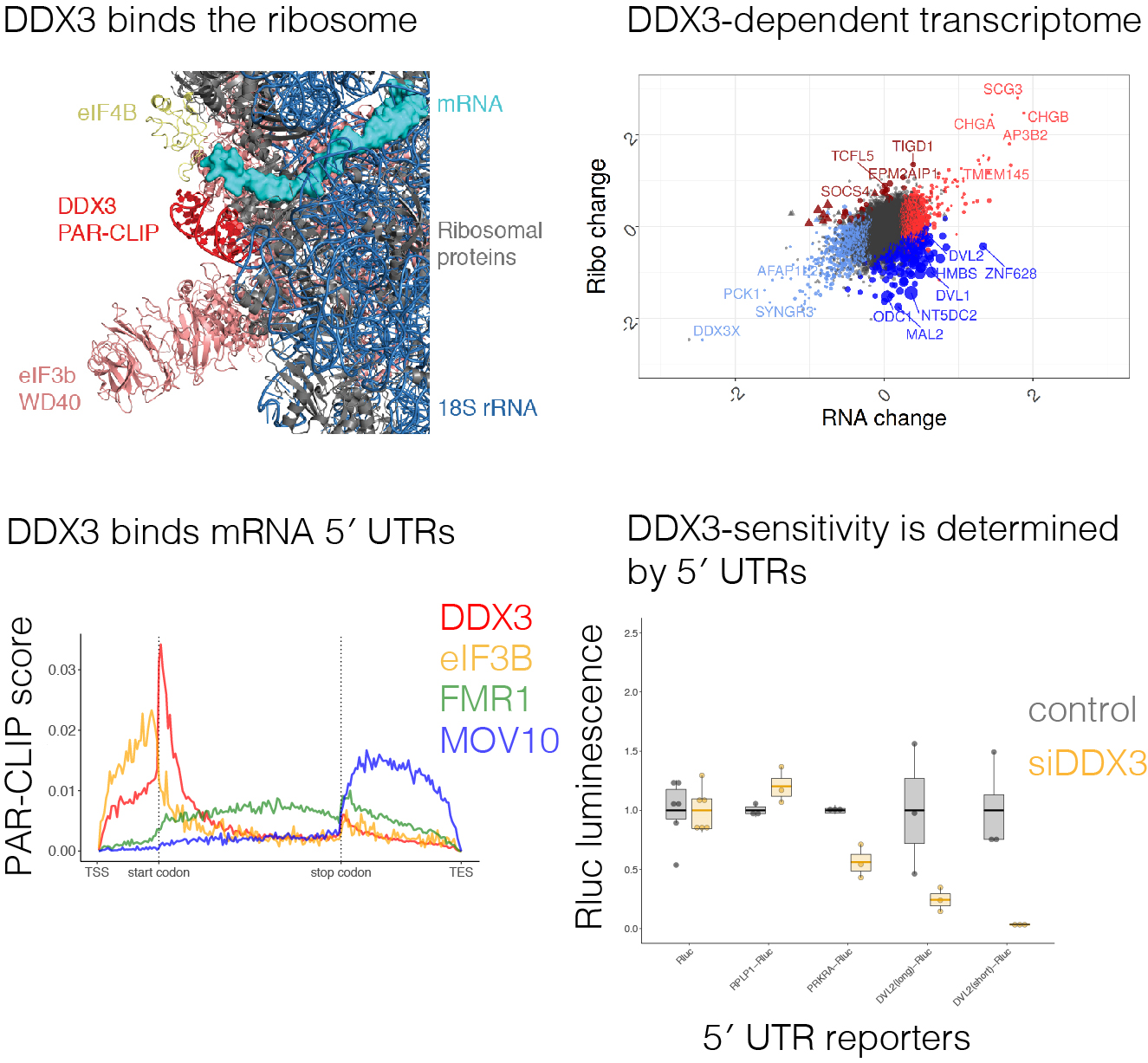 DDX3 depletion represses translation of mRNAs with complex 5′ UTRs
DDX3 depletion represses translation of mRNAs with complex 5′ UTRs
Calviello L, Venkataramanan S, et al., Landthaler M, and Floor SN. (2021)
 Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development.
Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development.
Lennox AL, Hoye ML, et al., Floor SN, Silver DL, and Sherr EH. (2020)
For a complete list of publications please click here or grab an RSS feed
Stephen can be reached at stephen@floorlab.org
The lab is closed as of 2024 and is not accepting new members.
Helen Vander Wende joins as a postdoc!
October 2022: Till Schroeder joins as a masters student!
September 2022: Sohyun Gu joins as a postdoc!
September 2022: Ziad is named a UCSF Discovery Fellow!
August 2022: Congrats to Albert for his F30 fellowship!
July 2022: Welcome to Hetvee Desai from UCSC joining for the summer!
September 2021: Welcome to Jess joining as a postdoctoral fellow!
September 2021: Congrats to Ziad for passing his quals!
July 2021: Welcome to Jesslyn joining as a graduate student and Angela joining as a technician!
June 2021: Stephen is a Pew Scholar in the Biomedical Sciences!
June 2021: José is awarded a NIGMS F31 fellowship!
May 2021: Welcome to Margaret Gadek, joining as a MSTP graduate student!
March 2021: Welcome to Sam Kwok, joining as a technician!
November 2020: Ziad joined as a BMS/MSTP graduate student!
September 2020: Two new preprints posted in the same week!
September 2020: Congratulations to José for being selected as a UCSF Discovery Fellow!
May 2020: Albert Xu joined as a BMS/MSTP graduate student!
May 2020: Congratulations to José for passing his qualifying exam!
August 2019: Welcome to Ann Deng, a new technician, and José Liboy who has joined the lab!
June 2019: Kevin is awarded a predoctoral fellowship from the TRDRP!
May 2019: Our preprint identifying the set of mRNAs that require DDX3 for translation is out!
May 2019: Congrats to Kevin, PhD candidate, for passing his qualifying exam!
December 2018: Welcome to Yizhu Lin, our new postdoc joining from Joel McManus's lab!
October 2018: The lab is funded by the NIH New Innovator program!
July 2018: Welcome to Lorenzo Calviello, our new postdoc joining us from Uwe Ohler's lab!
July 2018: Welcome to Luisa Tacca, a visiting graduate student from Jamie Cate's lab at UC Berkeley!
June 2018: Welcome to Kevin Wilkins, our first graduate student, and Katie Blackwell for the summer from SRTP!
March 2018: Welcome to Srivats Venkataramanan, our first postdoc from Tracy Johnson's lab!
February 2018: The lab is funded by the TRDRP for medulloblastoma research!
December 2017: The lab is funded by the UCSF PBBR!
July 2017: Bao and Malvika join in starting the lab!